structRNAfinder: an automated toolkit for the prediction and functional annotation of
non-coding RNAs through secondary structure inference
StructRNAfinder is an automated tool for the identification, functional annotation and taxonomic assignation of RNA families through secondary structure inference. It integrates third-part softwares to compare nucleotide sequences with covariance models, generate secondary structures and functional annotations in a stand-alone toolkit with friendly reports and outputs useful for downstream analysis and data exploration.
The tool is built using Perl, JavaScript and HTML. It integrates different tools (Infernal and RNAfold) in an optimized and streamlined way to facilitate the annotation of ncRNAs based on sequence/structure comparisons and databases (Rfam) cross-referencing.
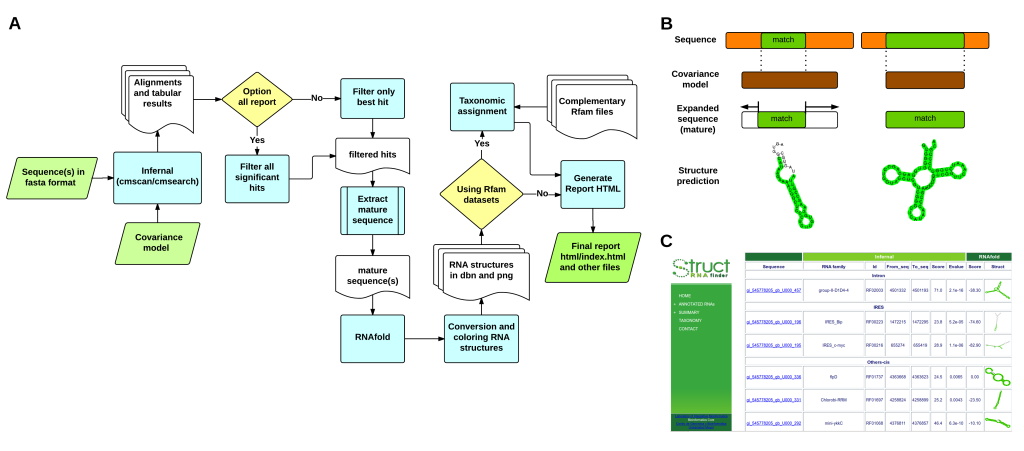
Figure 1 – (A) General workflow implemented in structRNAfinder. (B) Schematic view of the sequences/covariance models comparisons, mature sequence deduction and secondary structure prediction. The matching alignment between sequences and CMs is highlighted in green (C) Overview of one of the HTML reports provided.
Downloading and Installing
Download and collaborate on StructRNAfinder development by accessing it in our GitHub account. All information to install and run it can be accessed there. Any questions please feel free to contact as on the e-mail provided on the end of this page.
Exemplary Reports, Documentation and Tutorials
Exemplary reports can be accessed at: http://200.12.130.109/
Pre-Requisites
StructRNAfinder uses third-part and softwares in-house Perl scripts in order to perform all its workflow. All them are automatically installed and configured when installing the tool. Bellow a list of softwares necessary to use it.
– Perl
– Perl libraries libgd-perl and Bio::Graphics
– Infernal
– RNAfold
– Rfam covariance models
Contact
Please, feel free to contact us in case of comments, criticisms and suggestions at: viniciusmaracaja@integrativebioinformatics.me


